Date: Wed, 29 Mar 2017 13:13:45 +0000
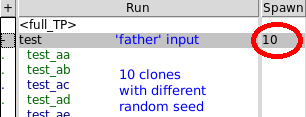
Czesc, You can find a Spawn number in the Run Tab, on the right side of the input name. Just click there and set the number. It will create several cloned inputs with different random seeds automatically. See attachment. Cheers, Wioletta ________________________________________ From: amucha_at_agh.edu.pl [amucha_at_agh.edu.pl] Sent: 29 March 2017 14:49 To: Wioletta Sandra Kozlowska Cc: fluka-discuss_at_fluka.org Subject: RE: [fluka-discuss]: merging outputs from different clones Czesc, thanks for the answer. Yes, indeed I can see all my files processing, but (following your example) they result in many .bnn files: test_aa_22.bnn test_ab_22.bnn etc I tried to change "Rules" of merging but it seems I still miss something and got a .bnn file for each clone separately. BTW - where can you set the "Spawn" number? I clone my inputs by "Run->Clone" sequence, then change seed manually. cheers, Agnieszka W dniu 2017-03-29 13:43, Wioletta Sandra Kozlowska napisał(a): > Czesc Agnieszka, > > First of all look into your Output Tab in flair. There you can check > how your files are processed and ensure if what you receive is the > merged file from all clones or the merged file from only one clone. > For example merging several clones should give you sth like this: > > Processing: test_22.bnn cmd=$FLUPRO/flutil/usbsuw >>>> test_aa001_fort.22 >>>> test_ab001_fort.22 >>>> test_ac001_fort.22 >>>> test_ad001_fort.22 >>>> test_ae001_fort.22 >>>> test_af001_fort.22 >>>> test_ag001_fort.22 >>>> ... > > Otherwise, you will see processing only one file. > > The easiest way is to create clone inputs using flair (setting 'Spawn' > number). Then flair will handle merging all clones after selecting > 'father' file. > > Moreover, if you have any problems, you can also use external > functions provided by fluka: usbsuw, ustsuw, usyswu, usxrea, usrsuw … > (you can find them in $FLUPRO/flutil). > > Cheers, > Wioletta > ________________________________________ > From: owner-fluka-discuss_at_mi.infn.it [owner-fluka-discuss_at_mi.infn.it] > on behalf of amucha_at_agh.edu.pl [amucha_at_agh.edu.pl] > Sent: 29 March 2017 10:44 > To: fluka-discuss_at_fluka.org > Subject: [fluka-discuss]: merging outputs from different clones > > Hi, > I want to speed up my simulations so I clone my input and use > multicore. > And I approached some problems with merging the results. This problem > has been already discussed: > http://www.fluka.org/web_archive/earchive/new-fluka-discuss/9612.html > but I still have doubts if I’m doing it correctly. > In Flair I highlightall clones inputs, then Process Data. I get outputs > like .bnn and .trk files that correspond and to one clone only (in the > filename there is > the clone number). > I’m almost 100% sure that If I choose “father” I get plots that come > also from one clone only – I can see this on plots and from statistics > in .tab files. Does anybody has any experience with this? > And the second question – how can I merge results from different clones > in the next session of Flair? I cannot highlight “father” input then > but > need to choose only one clone input. Is it possible to use Flair to do > this or do I need to process outputs with other tools or additional > scripts? > > Regards, > Agnieszka > > __________________________________________________________________________ > You can manage unsubscription from this mailing list at > https://www.fluka.org/fluka.php?id=acc_info > > > __________________________________________________________________________ > You can manage unsubscription from this mailing list at > https://www.fluka.org/fluka.php?id¬c_info
__________________________________________________________________________
You can manage unsubscription from this mailing list at https://www.fluka.org/fluka.php?id=acc_info
(image/png attachment: spawn.png)