Date: Thu, 2 Jul 2015 10:02:40 +0000
Hi Francesco,
could you send me your input file before the deletion of the preprocessor cards
to check.
Vasilis
________________________________
From: owner-fluka-discuss_at_mi.infn.it [owner-fluka-discuss_at_mi.infn.it] on behalf of Francesco Collamati [francesco.collamati_at_roma1.infn.it]
Sent: 01 July 2015 12:26
To: fluka-discuss_at_fluka.org
Subject: [fluka-discuss]: Flair preprocessor with Voxel geometry
Dear experts
I am using voxel geometry to import in FLUKA CT images of a mouse. Due to the high resolution of this image (~0.1mm^3 voxel), the whole CT results to have too many regions to be used in FLUKA, which gives the error **** Blank common exhausted in Pwxsst ****.
For this reason, I Thought to divide the CT in 3 “parts” along the Z axis, obtaining 3 separate voxel files.
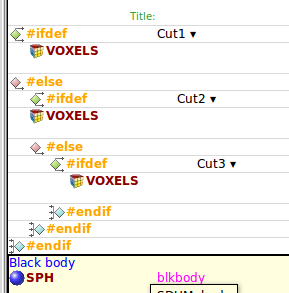
Now, I would like to be able to use the usual (and fantastic) preprocessor of flair with its defines and so on to create within the same .flair different runs selecting each time a different voxel file.
The system seems to work (see attachment), in fact I am able to see the different configurations by selecting them in the context menu from the geoviewer.
However, after a certain time, the program itself deletes all my #ifdef, #else and so on, leaving only one VOXEL card.
Is it due to the fact that the VOXEL geometry “doesn’t like” to be submitted to those conditions? Or is it just a bug?
As an alternative I tried to use 3 different .geo files, each one “calling” a different voxel file, but in this case it seems that flair wants an explicit presence of the VOXEL card, “extracting” it from the .geo file..
So, to sum up, is there any “handy” way to use a whole CT scan (I have had the same problem with a “whole body" human CT scan) in flair without having to create manually N different .inp files?
Thank you in advance
[cid:77C22A8B-B949-4D64-BD1C-230498BE8DA8_at_roma1.infn.it]
############################
Francesco Collamati Ph.D.
University of Rome Sapienza
INFN Rome
Enrico Fermi Building, room 203, tel. +39 06 49913453
francesco.collamati_at_roma1.infn.it<mailto:francesco.collamati_at_roma1.infn.it>
###########################
__________________________________________________________________________
You can manage unsubscription from this mailing list at https://www.fluka.org/fluka.php?id=acc_info
(image/png attachment: Schermata_2015-07-01_alle_11.51.01.png)