Date: Wed, 20 Apr 2016 07:56:39 +0000
Hi Khan,
yes you can calculate attenuation coefficients. There are several builtin scoring options in fluka
with that you can extract the flux and attenuation at every position, which you can convert it to
your attenuation coefficient. Please look the lectures on scoring on our basic course.
FLUKA runs on linux, however you can use fluka with Virtualbox on any machine windows, mac, etc.
Check the FLUPIX distribution in the download section of fluka.
Best
Vasilis
________________________________
From: owner-fluka-discuss_at_mi.infn.it [owner-fluka-discuss_at_mi.infn.it] on behalf of Drsalahuddin Khan [drskhan_at_ksu.edu.sa]
Sent: Tuesday, April 19, 2016 12:30
To: Roman Savinov; fluka-discuss_at_fluka.org
Subject: [fluka-discuss]: Attenuation coefficient measurement
Dear members,
I just join FLUKA but before getting ahead can anyone helpe me out with these questions, i highly appreciate that,
Whether FLuka code alone can calcualte attenuation coefficient of any material with gamma or neutorn soource as source?
is this code only work on LINUX system or if i do have Fortran language it may work?
thank you for your reply.
Regards,
Dr.Khan
________________________________
From: owner-fluka-discuss_at_mi.infn.it <owner-fluka-discuss_at_mi.infn.it> on behalf of Roman Savinov <rsavinov_at_calpoly.edu>
Sent: Tuesday, April 19, 2016 10:31 AM
To: fluka-discuss_at_fluka.org
Subject: [fluka-discuss]: Dose profile due to GCR
Dear FLUKA experts,
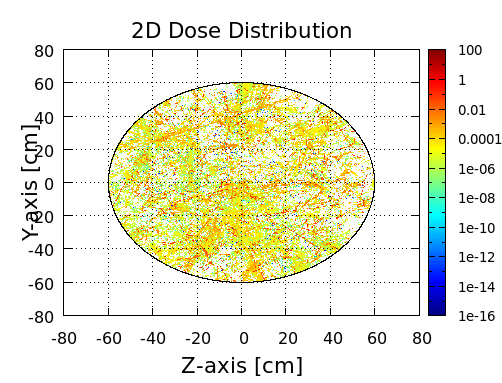
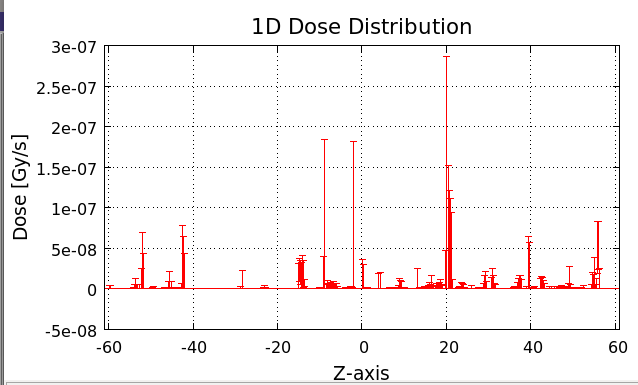
I have an emission sphere of GCR and a spherical water target. My expectation is that I should get a decreasing profile of dose distribution along the target radius (highest value on the surface, lowest toward the center). However, what I do get is a randomized dose distribution:
[cid:41dea6d6-3c88-4fcb-8e30-b5ff9e6dcda9]
[cid:c207fa74-bcca-42ad-813f-91510476a639]
Looks like 60 cm of water shows no attenuation whatsoever.
Can someone take a look at my input file and let me know what I do wrong?
I also include 6 GCR source files for the proton to carbon nuclei.
thank you,
Roman
________________________________
Disclaimer:
This communication is intended for the above named person and is confidential and / or legally privileged. Any opinion(s) expressed in this communication are not necessarily those of KSU (King Saud University). If it has come to you in error you must take no action based upon it, nor must you print it, copy it, forward it, or show it to anyone. Please delete and destroy the e-mail and any attachments and inform the sender immediately. Thank you.
KSU is not responsible for the political, religious, racial or partisan opinion in any correspondence conducted by its domain users. Therefore, any such opinion expressed, whether explicitly or implicitly, in any said correspondence is not to be interpreted as that of KSU.
KSU may monitor all incoming and outgoing e-mails in line with KSU business practice. Although KSU has taken steps to ensure that e-mails and attachments are free from any virus, we advise that, in keeping with best business practice, the recipient must ensure they are actually virus free.
__________________________________________________________________________
You can manage unsubscription from this mailing list at https://www.fluka.org/fluka.php?id=acc_info
(image/png attachment: pastedImage.png)
(image/png attachment: 02-pastedImage.png)