Date: Tue, 12 May 2020 23:23:10 +0530 (IST)
Dear Experts,
Please refer to this (asked in last mail): For DOSE-EQ scoring,
About mSv/h: after getting results in pSv/particle, i need to multiply
>> it by (3.7E+10 * 3600) to convert it to pSv/h. And later to mSv/h.
>> Hope I am doing it correct. Please comment.
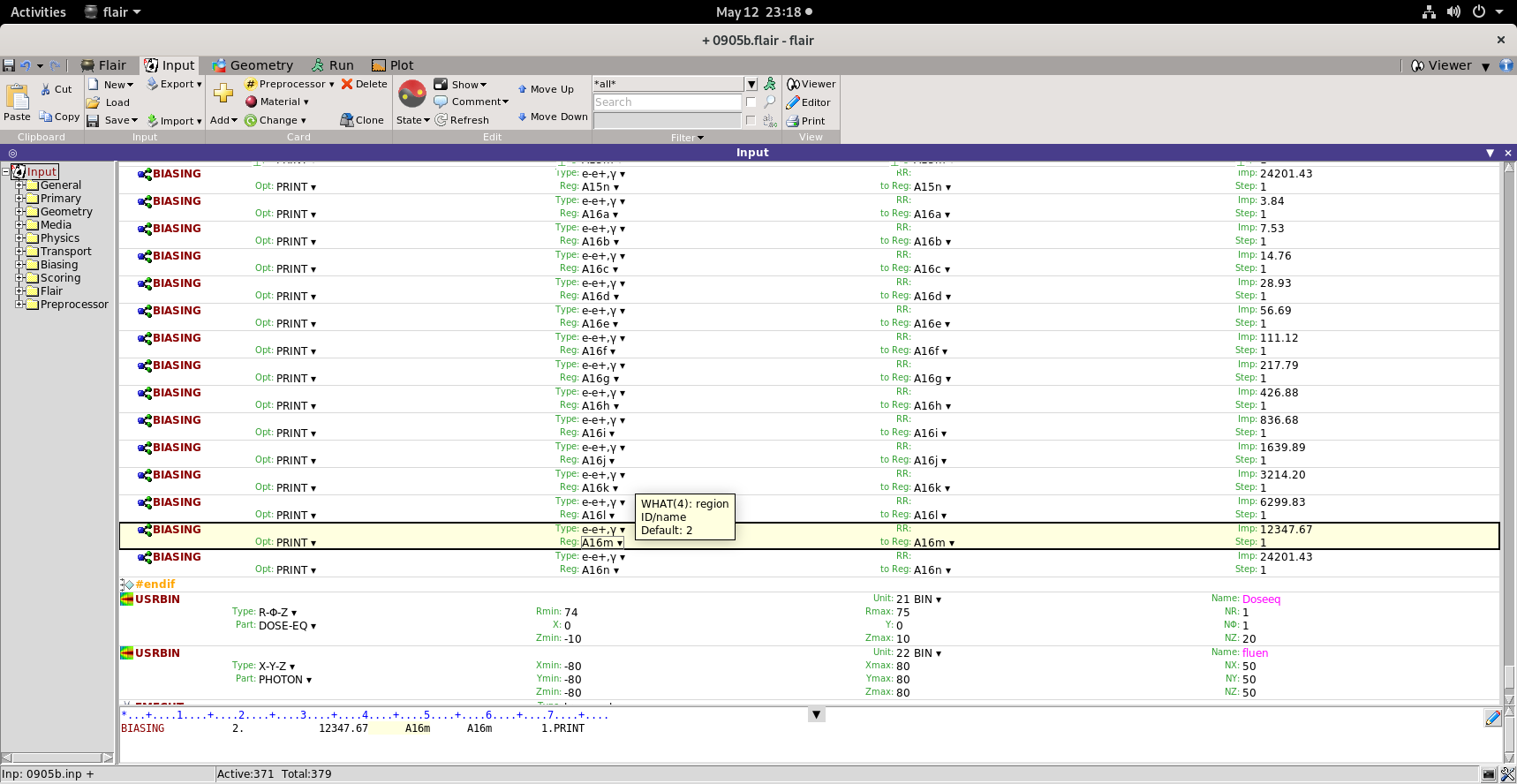
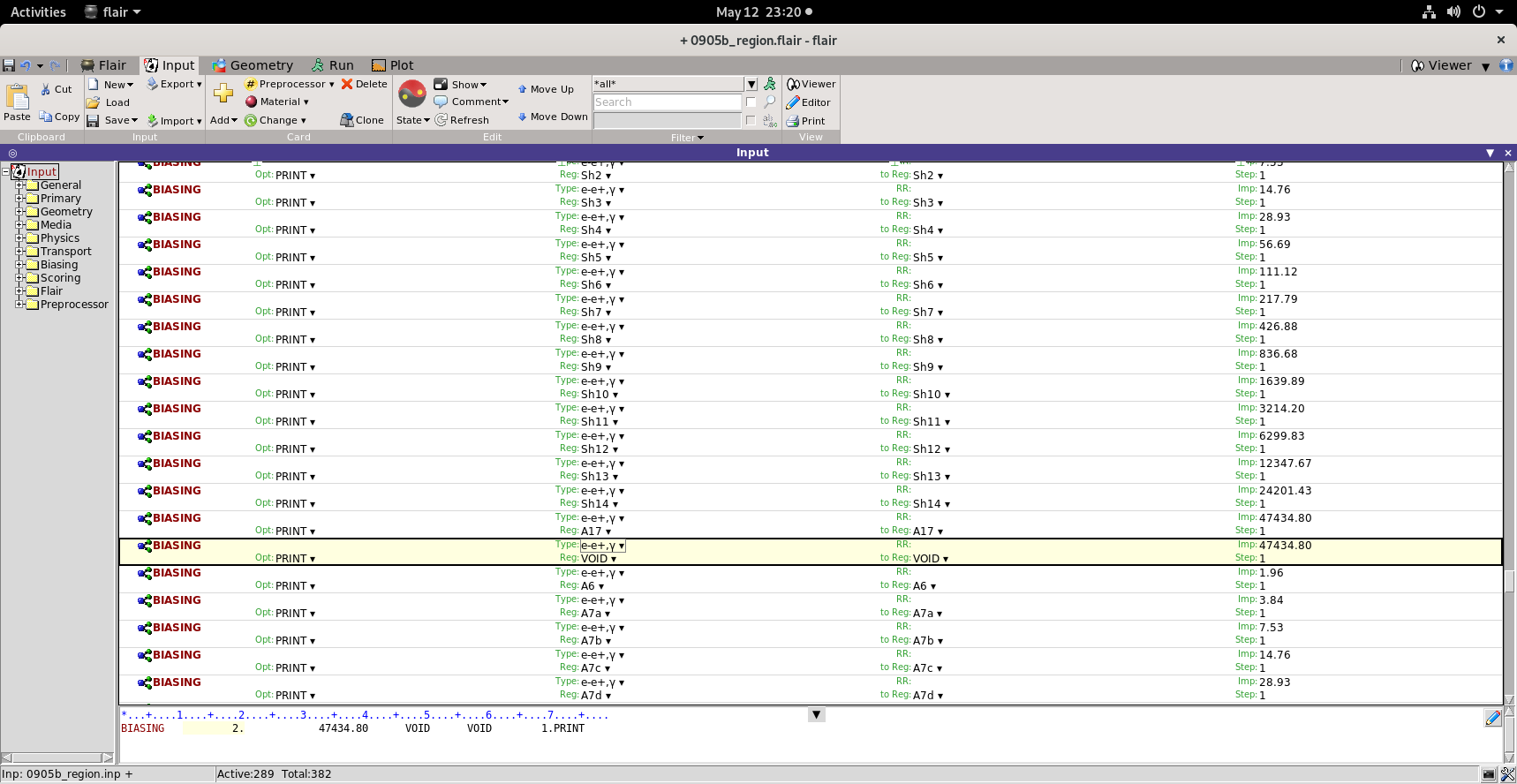
Secondly, I have attached the corrected Biased input, with the green regions biased and importance to outside air (VOID) given as same as the importance given to last region.
Even with this input, my results are
R - Z binning n. 1 "Doseeq " , generalized particle n. 240
R coordinate: from 7.4000E+01 to 7.5000E+01 cm, 1 bins ( 1.0000E+00 cm wide)
Z coordinate: from -1.0000E+01 to 1.0000E+01 cm, 20 bins ( 1.0000E+00 cm wide)
axis coordinates: X = 0.0000E+00, Y = 0.0000E+00 cm
Data follow in a matrix A(ir,iz), format (1(5x,1p,10(1x,e11.4)))
accurate deposition along the tracks requested
this is a track-length binning
2.3803E-09 2.2997E-09 2.2466E-09 2.2076E-09 2.2015E-09 2.1618E-09 2.0683E-09 2.0246E-09 2.0327E-09 1.9877E-09
1.9922E-09 1.9436E-09 1.9155E-09 1.9019E-09 1.9583E-09 1.9520E-09 1.8344E-09 1.7945E-09 1.8017E-09 1.8284E-09
Percentage errors follow in a matrix A(ir,iz), format (1(5x,1p,10(1x,e11.4)))
2.8653E+00 3.0830E+00 2.9042E+00 3.4062E+00 3.2796E+00 3.7762E+00 4.2888E+00 4.4818E+00 4.1917E+00 3.9561E+00
3.0624E+00 3.0086E+00 2.6655E+00 3.6244E+00 2.9617E+00 2.2595E+00 3.3564E+00 3.7204E+00 4.6390E+00 4.8849E+00
The order of the Dose-Eq is almost 1000 times higher than expected. That is the values which I expect in mSv/h, I am getting in Sv/h (after the conversion for Ci and hour).
Kindly comment and suggest.
Thanks in advance, regards,
Raksha Rajput.
----- Original Message -----
From: "paola sala" <paola.sala_at_mi.infn.it>
To: "tamer tolba" <tamer.tolba_at_uni-hamburg.de>
Cc: fluka-discuss_at_fluka.org
Sent: Saturday, May 9, 2020 8:37:56 PM
Subject: Re: [fluka-discuss]: Ques on Biasing
Hello
First a general answer:
All what is set through Irrprofi, Dcytimes, dcyscore, regards the
RADIOACTIVE DECAY of nuclei produced during irradiation, only. Prompt
radiations (for instance neutrons/photons produced in nuclear
interactions) are not affected by the irrprofi etc cards.
The special DCYSCORE -1 flag can be applied ONLY if raddecay is asked in
semi-analogue mode: it is used to score together prompt and delayed
radiation. It is not compatible with IRRPROFI
What is the difference between RADDECAY 2 (semi-analogue) and RADDECAY 1
(standard) way?
With raddecay 1, the activity for aech isotope at a given cooling time is
calculated analitically starting from the production of initial
radioisotopes in the prompt part, plus Bateman equations for the buildup
and decay through radiactive chains. therefore, it is possible to get
results for many cooling times in the same setup.
For raddecay 2, instead, the decay is treated "as real", in a Monte-Carlo
way: each decay time and channel is chosen randomly, until the chain ends
on a stable nucleus. In this way, there is no possibility to preset a
given cooling time. The user can only choose wether to score both prompt
ans radioactive, or only prompt (or set time intervals in Usryield or
time intervals in scoring routines)
detailed answers below
> Hi,
>
> May I pop-up here, as I have very a close/related question to Raksha
> last question!
>
> For the Eq-Dose (and Activity) estimations, assuming irradiation profile
> of: beam intensity (PpS), Irradiation time (t_Irr) and cooling/decay
> time (t_Col):
>
> - For EQ-DOSE during exposure: I put t_Col = -1, right?
No. If you wish to have the "prompt" dose, I mean from the primary beam
and the particles produced immediately by the beam, you simply do not
associate any cooling time to your detector. It will contain only the
promot radiation
>And the result
> will be in PSv/Primary, right?
yes
> And to get it to PSv/s I need to divide
> by t_Irr and multiply by PpS, right?
No. You have to multiply by PpS, only.
>
> - For EQ-DOSE after decay: I put t_Col > 0 [s], right? And the results
> will be in PSv/s, right? And to get it in pSv/h I need to multiply by
> (3600), right?
Yes
>
> - For EQ-Dose prompt: Should I put t_Col = 0? if yes, the results will
> be in PSv/primary? if yes, how to convert it to PSv/s? Should I multiply
> it by PpS and divide by t-Irr?
>
If you want the EQ-Dose produced by radioactive nuclei at the end of
irradiation, use t_Col=0 and get it in PSv/s. If you wish to get it at a
given time during irradiation, use a negative t_Col (conunting back from
the end of irradiation) and get it in pSv/s
> Regards,
>
> Tamer
>
> On 08.05.2020 07:58, Raksha Rajput wrote:
>> Dear Experts,
>>
>> Thanks alot for the explanation and the corrections suggested.
>>
>> About mSv/h: after getting results in pSv/particle, i need to multiply
>> it by (3.7E+10 * 3600) to convert it to pSv/h. And later to mSv/h.
>> Hope I am doing it correct.
>>
>> Regards,
>> Raksha R.
>>
>> ----- Original Message -----
>> From: "paola sala" <paola.sala_at_mi.infn.it>
>> To: "Raksha Rajput" <rakshak_at_britatom.gov.in>
>> Cc: answers_at_pcfluka.mi.infn.it, fluka-discuss_at_fluka.org
>> Sent: Thursday, May 7, 2020 6:51:35 PM
>> Subject: Re: [fluka-discuss]: Ques on Biasing
>>
>> Hello
>> answres below
>>> Dear Experts,
>>> Thank you for your time.
>>>
>>> Please find attached the biasing I have used in the input. As I
>>> understood
>>> from one of the examples given in the FLUKA Course, giving "BIASING
>>> 0.0 1. BLKBODY _at_LASTREG 1.PRINT" means giving importance 1
>>> to
>>> regions outside shield.
>> It means giving importance 1 to ALL regions, it is a sort of
>> initialization.
>> Adding more BIASING cards after this one will change the impostance of
>> the
>> various regions.
>> However, if you leave the outside air to 1, almost all particles will be
>> killed when they go from the shield (high impos=rtance) to the outside
>> air
>> (low importance).. thus all the biasing will be lost there. The outside
>> air should have the same importance as the last layer of the shield.
>>
>>> Am I wrong to understand this ? I am sorry, I
>>> missed to have added any biasing for green regions (which are SS
>>> material).
>> Same as before.
>>
>>> Secondly, by the results are higher by a factor of 1000, I mean to say,
>>> the dose rates which I should get in mSv/h, I am getting in Sv/h.
>> ? results for DOSE-EQ are in picoSievert/primary for prompt radiation,
>> picoSievert/second for specific decay times.
>>
>>> Thanks and regards,
>>> Raksha R.
>>>
>>>
>>> ----- Original Message -----
>>> From: answers_at_pcfluka.mi.infn.it
>>> To: "Raksha Rajput" <rakshak_at_britatom.gov.in>
>>> Cc: fluka-discuss_at_fluka.org
>>> Sent: Wednesday, May 6, 2020 8:41:57 PM
>>> Subject: Re: [fluka-discuss]: Ques on Biasing
>>>
>>> Hello
>>> The geometry looks fine, I cannot judge on biasing not knowing the
>>> factors
>>> you used. However: what is the weight assigned to the green regions in
>>> your plots? And to outside air?
>>> Could you please explain what do you mean by "results a factor 1000
>>> higher than expected"?
>>> Thanks
>>> Paola
>>>
>>> On Tue, 28 Apr 2020, Raksha Rajput wrote:
>>>
>>>> Dear Experts
>>>>
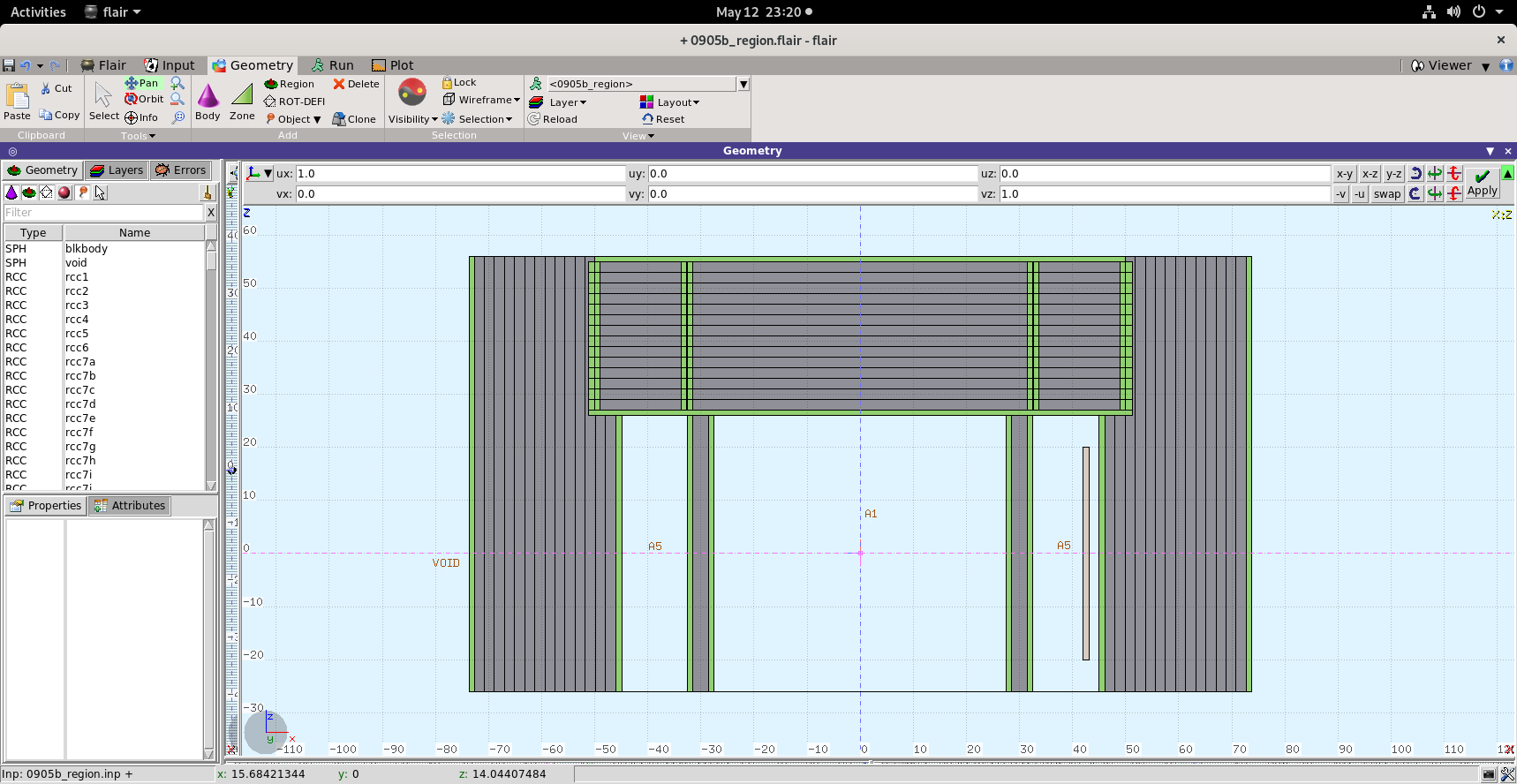
>>>> Please find attached the geometry cross-section.
>>>> 1. RCC (A10) is cut by planes to divide it into regions, for Biasing
>>>> (surface-splitting, increasing no. of photons per region)
>>>> 2. RCC(A14) is also cut by same planes (as these are infinite), to
>>>> divide for biasing. Is this the correct way ? OR, it should be done by
>>>> making concentric RCCS as is done for RCC(A7). A10 & A14 for Axial
>>>> points (Top of the shield).
>>>> 3. RCC(A7) is divided into regions by making concentric RCCs - to
>>>> calculate the dose rates in radial points.
>>>> Kindly suggest whether the regions are divided correctly.
>>>> Or, please suggest corrections.
>>>> The results I am getting are higher(by a factor of 1000) than
>>>> expected.
>>>>
>>>> Thanks in advance.
>>>> Regards,
>>>> Raksha.
>>>>
>>>>
>>>>
>>
>> Paola Sala
>> INFN Milano
>> tel. Milano +39-0250317374
>> tel. CERN +41-227679148
>>
>>
>> __________________________________________________________________________
>> You can manage unsubscription from this mailing list at
>> https://www.fluka.org/fluka.php?id=acc_info
>>
> --
> Dr. Tamer Tolba
> Institut für Experimentalphysik,
> Universität Hamburg ,
> Luruper Chaussee 149,
> 22761 Hamburg - Germany
>
> Tel.: +49 (0)40-8998-4872
>
> __________________________________________________________________________
> You can manage unsubscription from this mailing list at
> https://www.fluka.org/fluka.php?id=acc_info
>
Paola Sala
INFN Milano
tel. Milano +39-0250317374
tel. CERN +41-227679148
__________________________________________________________________________
You can manage unsubscription from this mailing list at https://www.fluka.org/fluka.php?id=acc_info
__________________________________________________________________________
You can manage unsubscription from this mailing list at https://www.fluka.org/fluka.php?id=acc_info
(image/png attachment: RZbinning.png)
(image/png attachment: imp_VOID.png)
(image/png attachment: corrected_input.png)